In drug discovery, the journey from identifying a promising compound to an approved drug is fraught with challenges. A staggering 90% of potential drugs fail to reach patients. This high failure rate often stems from misidentifying the right protein targets in the human body that a drug can effectively bind to and act upon. Traditional methods for finding these 'druggable' proteins combine experimental approaches, such as Nuclear Magnetic Resonance, with computational methods that analyse protein features. These methods are often slow, imprecise, and limited, resulting in substantial financial and time investments. The recent rise of machine learning and Artificial intelligence tools has now exponentially expedited this process, leading to new drug discoveries in rapid times.
In another boost to this process, a team of researchers from the Indian Institute of Technology (IIT) Bombay has developed an innovative new tool, DrugProtAI. Their new computational framework is designed to predict whether a protein can be effectively targeted by a drug, offering a powerful new ally in the fight against diseases.
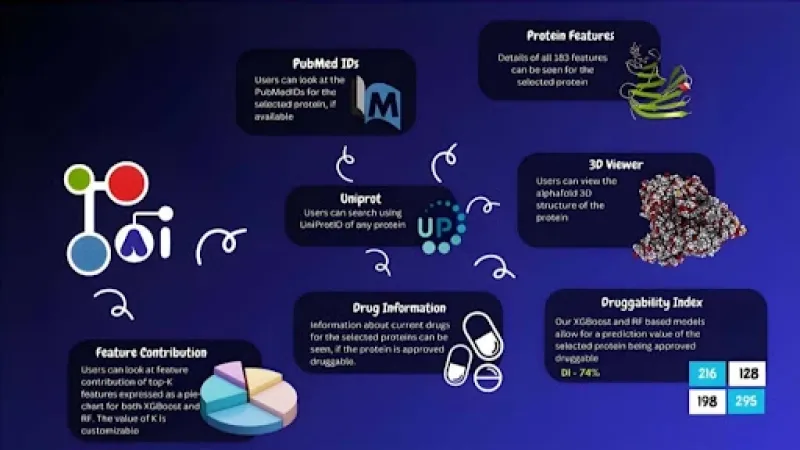
DrugProtAI's novelty lies in its ability to analyse proteins using a vast array of information, far more than previous tools. Instead of just looking at a protein's basic building blocks, amino acids, DrugProtAI considers 183 different characteristics. It examines a protein's physical and chemical properties, its sequence (the order of its amino acids), how it interacts with other proteins, its location within a cell, and how it's modified after it's made. Their comprehensive approach, which draws data from major biological databases such as UniProt, DrugBank, PubMed, and 3D protein structure predictions from AlphaFold, creates a much richer picture of each protein.
Did you know that only about 10% of all potential drug candidates successfully make it through clinical trials to become approved medicines? The human body contains over 20,000 different proteins, but only a fraction of these are considered 'druggable' targets for medicines. |
One of the biggest challenges in developing such a tool is dealing with imbalanced data. In the human body, many more proteins aren't 'druggable' than those that are. If an AI model is trained on such skewed data, it may become biased and struggle to identify the rare, yet crucial, druggable proteins accurately. DrugProtAI addresses this challenge directly with a strategy known as a 'partitioning-based ensemble method.' The vast pool of non-druggable proteins is divided into smaller, more manageable groups. Then, multiple AI models are trained, each learning from one of these smaller groups in addition to the complete set of druggable proteins. This ensures that the models get a balanced view, preventing them from overlooking essential patterns. The researchers found that two popular machine learning algorithms, Random Forest and XGBoost, performed exceptionally well within this framework, achieving a median accuracy of 87% in predicting drug targets.
To ensure their tool was truly robust, the team conducted a rigorous blind validation test on DrugProtAI. They used it to predict the druggability of proteins that had only recently been approved as drug targets. These were proteins the AI had never encountered during its training. The results showed that DrugProtAI correctly identified 61 out of 81 newly approved drug targets, demonstrating its real-world applicability and superior performance compared to existing tools, like SPIDER and DrugTar.
Beyond making predictions, DrugProtAI also helps researchers understand why a protein is considered druggable. Using a technique called SHAP (SHapley Additive exPlanations), the tool identifies the key features that contribute most to a protein's druggability. For instance, the presence of kinases (a type of protein often involved in cell signalling), specific secondary structures, and a protein's 'instability index' were found to be strong indicators. This interpretability is vital because it provides biological insights, enabling researchers to make informed decisions rather than relying solely on a black-box prediction. While the team also explored deep learning methods that sometimes yielded slightly higher prediction scores, they noted that these methods could not often explain why they made a particular prediction, making DrugProtAI's interpretable approach particularly valuable.
DrugProtAI could streamline the identification of promising drug targets and significantly reduce the time and resources required for developing new medicines. The tool is also made freely available online, making this technology accessible to researchers worldwide. By providing a clear, unbiased, and accurate method for assessing protein druggability, DrugProtAI is poised to accelerate the development of life-saving drugs, bringing us closer to a future where more effective treatments are available for a broader range of diseases.
This article was written with the help of generative AI and edited by an editor at Research Matters.