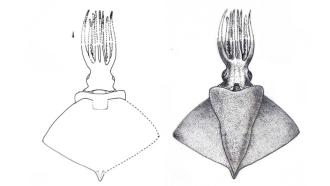
Image: Hydra vulgaris with suppressed TMOD1 expression showing the non-detaching Y-shaped bud still attached to the parent Hydra. Credit: Authors, https://doi.org/10.1016/j.isci.2025.113759
The tiny freshwater polyp Hydra vulgaris has captivated biologists with its astonishing ability to regenerate and reproduce asexually. One of its most fantastic party tricks is that when a hydra is cut into segments, each of the segments can grow into an individual Hydra, essentially making it biologically immortal. However, the precise molecular mechanism that governs the physical separation of a mature offspring (or bud) from its parent has remained surprisingly elusive. Now, a team of researchers from Agharkar Research Institute (ARI), Pune, has solved this long-standing developmental mystery, revealing an evolutionarily conserved mechanism with profound implications for human health.
In their new study, the researchers have pinpointed the actin-binding protein Tropomodulin-1 (TMOD1) as the essential molecular switch for the final stage of Hydra reproduction. They demonstrate that TMOD1 is absolutely necessary for forming the basal constriction, which is the critical pinching-off process that allows the offspring polyp to completely detach from the parent and begin its independent life.
Did You Know? Hydra are related to jellyfish and sea anemones! They are classified as cnidarians, a group of animals that are more ancient than insects and vertebrates. |
The evidence of TMOD1's vital role came from the team's loss-of-function experiments. Using a gene-silencing (suppression or blocking of a gene) technique called siRNA-mediated knockdown, the researchers suppressed TMOD1 expression in budding Hydra. While the parent polyps and their buds survived, the crucial step of basal constriction formation was dramatically inhibited. Instead of separating, the fully grown offspring remained fused to the parent body for weeks, creating a distinctive, non-detaching Y-shaped Hydra.
Further investigation into these compromised polyps showed that the internal cellular skeleton, specifically the F-actin network, lacked the organised stress fibres and proper cell shape required to form the final constriction ring. This confirmed TMOD1’s function in stabilising and organising the F-actin network within the ectodermal cells (the outer layer of an early embryo) at the parent-offspring junction. This stability enables the coordinated cellular contraction needed for detachment.
The researchers also uncovered the upstream signalling mechanism that controls the TMOD1 protein’s activity. The team established that TMOD1 expression is strongly regulated by the Fibroblast Growth Factor Receptor-Extracellular Signal-Regulated Kinase (FGFR-ERK) signalling pathway. By applying pharmacological inhibitors to block FGFR signalling and ERK phosphorylation, they found that TMOD1 expression was almost entirely suppressed at the crucial detachment site. This demonstrates that FGFR-ERK signalling activates TMOD1 in the ectodermal cells at the parent-bud junction, which then proceeds to organise the F-actin network for the physical constriction and separation of the bud.
While it was long known that bud detachment was achieved through an F-actin-based constriction at the parent-offspring junction, the precise molecular switch regulating this F-actin formation was previously considered elusive. Earlier studies had shown that the FGFR signalling pathway was crucial for the overall detachment process, as inhibition of the FGFR-like gene in Hydra (called kringelchen) stalled bud separation. However, the downstream effector linking FGFR-ERK signalling to the physical organisation of F-actin was a missing link. This new study fills that gap by establishing the direct relationship: the FGFR-ERK signalling axis regulates the expression of the actin-binding protein TMOD1, thereby providing the complete molecular machinery necessary for tissue separation in Hydra.
The long-term impact of this discovery extends far beyond the humble Hydra. TMOD1 is a protein highly conserved across evolution, having been maintained from primitive cnidarians to humans. In higher vertebrates, mutations in the TMOD1 gene are directly linked to severe inherited heart conditions, specifically a form of childhood cardiomyopathy (disease of the heart muscle).
Cardiomyopathy often involves the elongation of thin filaments in heart muscle cells, which affects the heart's ability to contract effectively. In higher animals, if TMOD1 activity is compromised, complex compensatory mechanisms often activate to mask the full extent of the defect, making it difficult for researchers to study the protein's pure functional loss. Since the Hydra is much simpler compared to humans, its straightforward reliance on TMOD1 for tissue separation provides a simpler, clearer model for molecular analysis. By revealing how the TMOD1 protein and its regulatory pathway (FGFR-ERK) operate in Hydra, researchers have established a foundational model directly applicable to human cellular biology.
This article was written with the help of generative AI and edited by an editor at Research Matters.
The article was edited to correct an inaccuracy and to correct the location. The error is regretted.